When things are too small to see with a naked eye, as is the case for many biological systems, you use a microscope. But a lot of highly interesting biological structures are so small that even a microscope cannot resolve them, at least not a standard light microscope. That is because light itself has a certain size. Things that are smaller than half the wavelength of the light used for imaging cannot be resolved, and since the smallest wavelength of visible (blue) light is about 400 nm, standard light microscopy cannot resolve the details of structures below 200 nm. However, several techniques have been developed to get around this limit.
One trick is to image individual fluorescent molecules, for example by using specialized fluorescent proteins that can be switched to blink on command. Each individual little point of light that the microscope detects can then be resolved no better than a normal light microscope can manage. But because we know the source of the light is a single molecule, the exact position of the fluorophore can be traced back to a much higher accuracy. This technique is called single-molecule localisation microscopy.
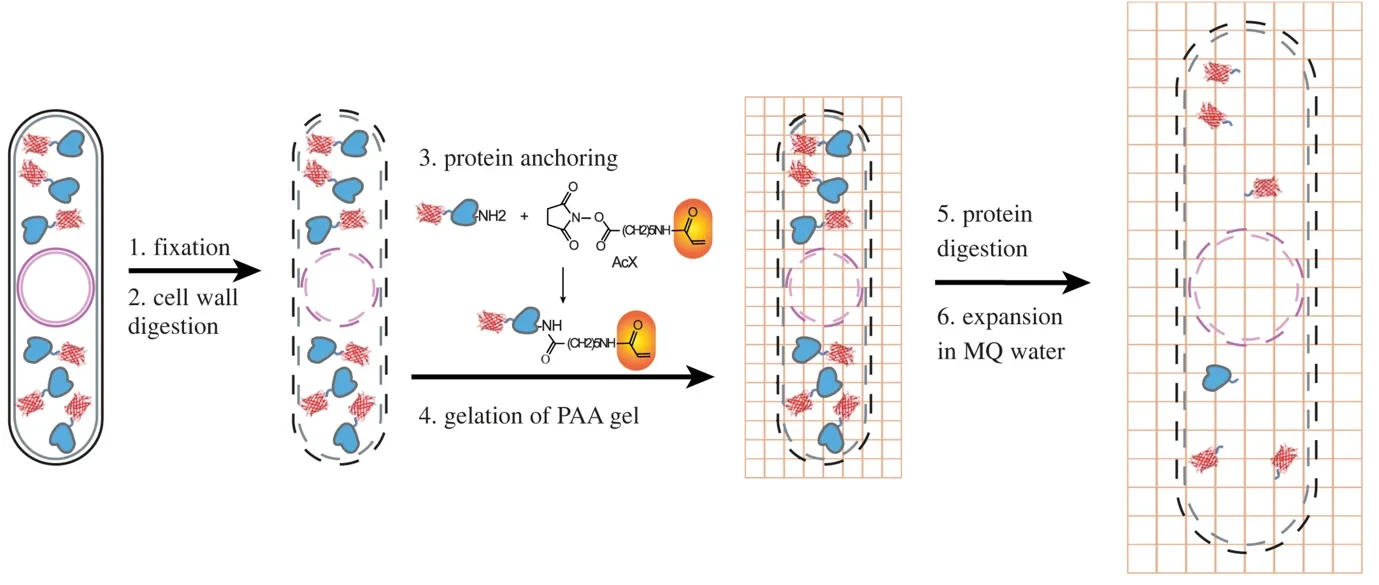
Another trick is to simply expand your sample. Again, fluorescent molecules are used here, in combination with some special plastic polymers (which, by the way, also play a key role in diapers keeping your baby dry). The fluorophores are glued to the polymer in the location they are at within a cell, then the cell itself is destroyed and – adding water - the polymer is expanded and the structural details – now about five times larger – become visible. Each technique by itself is a powerful way of learning about systems much smaller than usually visible under the microscope, but combining the two allows diving even deeper into small structures than could not be resolved by either technique alone.
In our new article, we show how Single molecule and Expansion Microscopy can be combined to look at some of the rarer proteins in fission Yeast – a SExY protocol indeed. To do that, we needed to overcome a number of obstacles. For example, it turns out that our yeast cells have some very hardy cell wall ‘belts’ that resisted digestion and restricted their expansion, giving rise to hourglass-shaped cells – SExY perhaps, but not entirely useful from a scientific point of view! Furthermore, creating the polymer and destroying the cell to allow expansion does not leave the fluorescent proteins unscathed, so we optimized various aspects of the protocol to push survival, the so-called retention yield of the fluorophores up to ca. 50%. Notable innovations included the addition of a radical quencher when creating the polymer, reducing the burden of reactive chemicals present in this step that were damaging our fluorescent reporters. In its optimized form, the protocol can now be used to visualize protein structures at the nanostructural level – even for proteins that exist at low abundance, such as nuclear proteins cbp1 and mis16, and the centromere-specific histone protein cnp1.
Started 2016 in Marburg, this project moved along with the authors to be worked on in Pittsburgh and Paris, and was eventually finalized here in Bonn at the IfMB. We are happy to announce that it has now been published in the Royal Society’s Open Biology Journal.